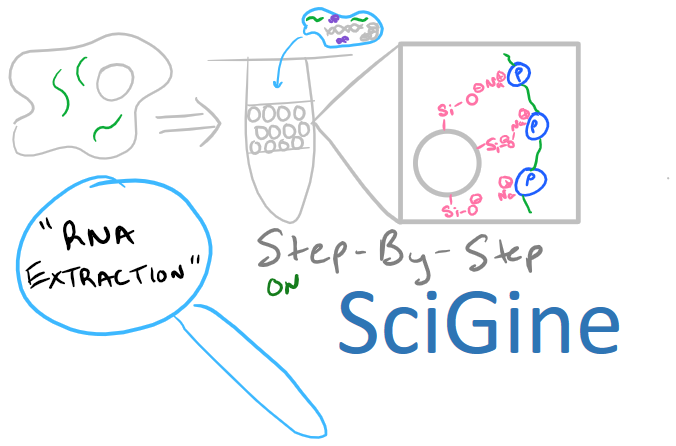
RNA Extraction: General Summary
RNA extraction and isolation is a precursor for many methods in molecular biology including Northern Blotting, RT-PCR, and Microarray analysis. This blog post will focus on this precursor method as opposed to other biology techniques on the Scigine blog where I’ve focused on direct analytical techniques. Nonetheless, there is a lot to learn about RNA isolation and plenty of theory that might be applicable in your research.
Note: If you’re writing research papers, I highly recommend Grammarly – it’s a free grammar check plugin for Chrome. Try it out here…
Important aspects related to RNA Extraction
While everyone knows DNA is double stranded and helical, few people know that RNA is typically a single stranded polymer. However, secondary structures do emerge within RNA due to complementary base pairing and structures such as tRNA can be double stranded. Also, due to secondary structures, flexible regions of RNA can actually catalyze the cleavage of phosphodiester linkages in nearby RNA chains.
The base polymer looks like this:
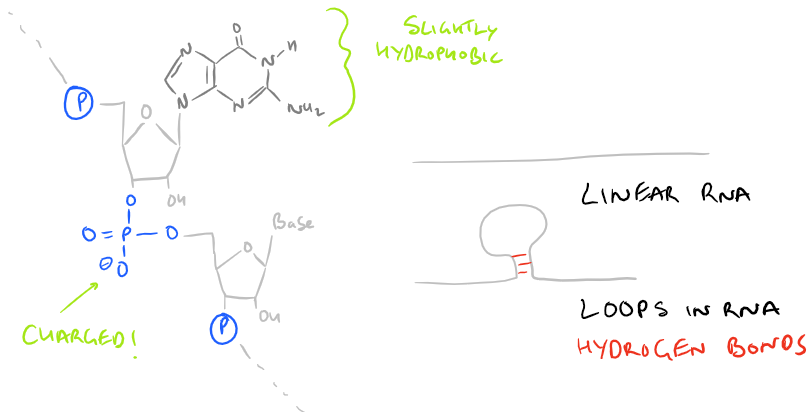
The RNA polymer has several ionic groups, inter-chain hydrogen bonds, and it is extremely hydrophilic. All of these forces need to be overcome in order to purify RNA from DNA, carbohydrates, and lipids which have similar structures and physical properties. Polymers of RNA can be short or long, but generally smaller strands that are less than 100 nu cleotide bases are hard to purify because they don’t separate well. Additionally, there are several RNAses present in cells and tissues that can catalyze cleavage of RNA chains, so extreme care needs to be taken to prevent degradation of RNA during RNA isolation and purification procedures.
RNA Extraction Method Guide
Typically RNA Extraction procedures start with cell lysis. A buffer that includes Guanidine Thiocyanate or other chaotropes is necessary to mask charges on RNA and water so that the polymer can be purified using solid-phase techniques. Chaotropes linearize the RNA polymer, disrupt hydrogen bonding and destroy the activity of any RNAses present in the cell lysate. Due to their ability to disrupt hydrogen bonding, they also lyse cells by disbanding the phospholipid bilayer.
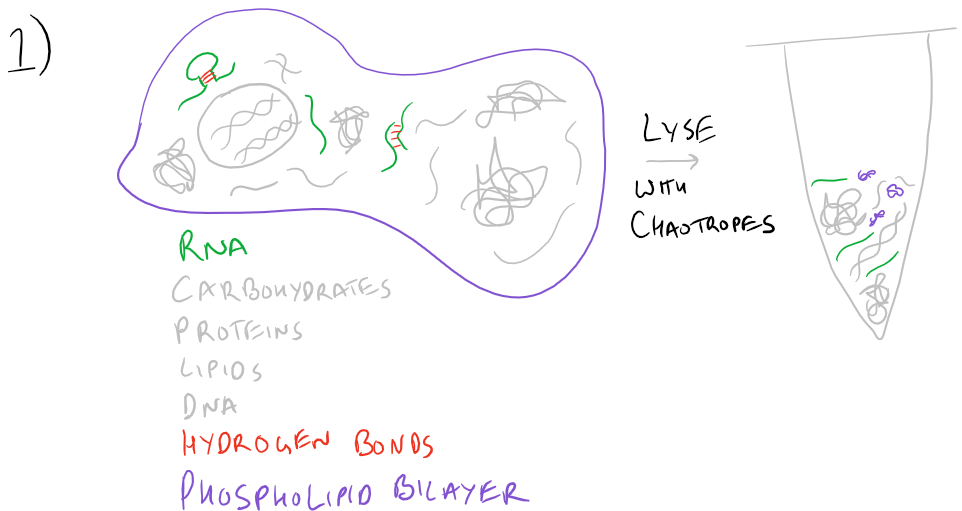
To facilitate homogenization, a homogenization column (imaged below) may be used. By passing at high speeds through resins, viscous polymers such as DNA and lipids can be separated and the cell lysate will flow more easily.
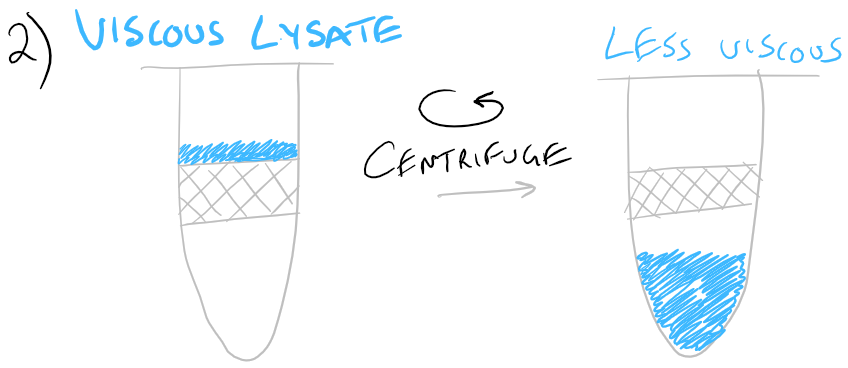
Ethanol is then added to: reduce the overall water concentration in the sample and precipitate proteins. As you would expect, RNA is highly soluble in water. A spin column (solid-phase support, imaged below) is then used to bind the RNA from the cell lysate.
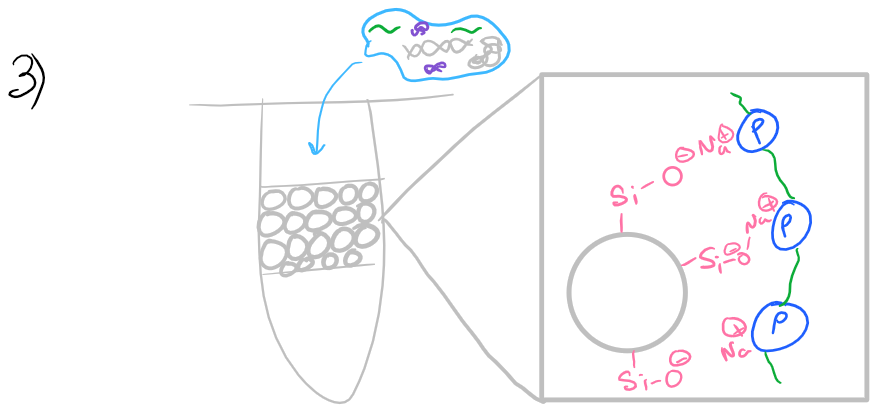
Initially a low chaotrope concentration wash buffer is used to clean the RNA sample and remove proteins while RNA remains attached to the column.
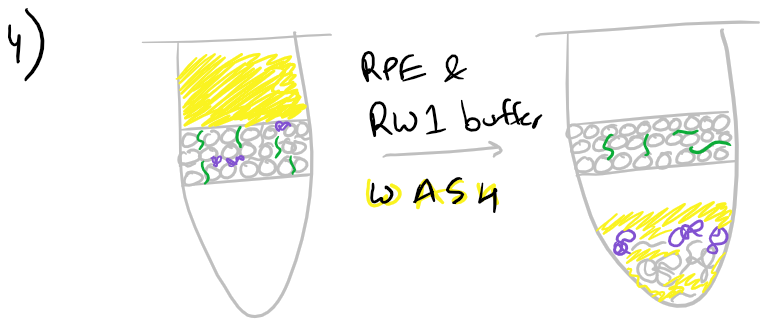
Next, an ethanol wash removes some of the chaotropic salts that were left over from previous washes.
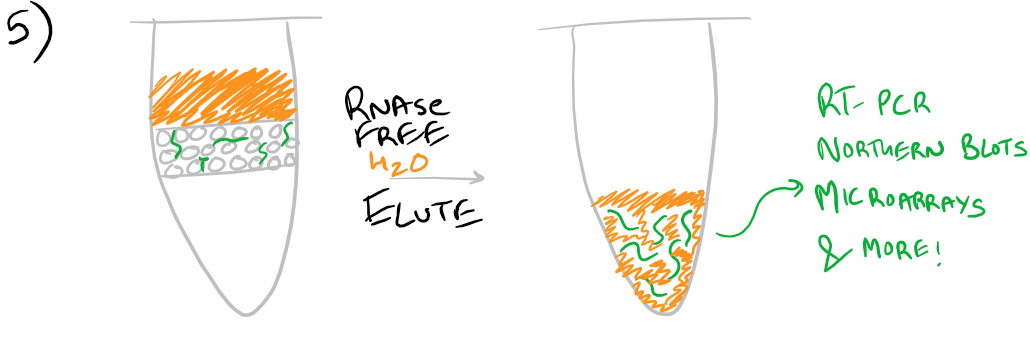
Finally, without chaotropic salts present, RNAse-free water can be used to elute the RNA sample from the column
Note: Grammarly is a free grammar check plugin for Chrome. I used it for this article and really like it! Try it out here…
RNA Extraction and Purification: Step by Step
Here’s a technique for RNA Isolation and cDNA preparation from Pancreatic Islets. We can use this technique for analysis of gene expression later on. Our strategy is to use solid-phase extraction of nucleic acids from complex cell lysate samples and then we can prepare the cDNA to examine the expression of genes such as insulin.
Materials for RNA Extraction and cDNA preparation:
Isolated pancreatic islets
RNeasy minikit (#74104 Qiagen)
QIAShredder columns (#79654 Qiagen)
Omniscript reverse transcription kit (#205110 Qiagen)
Oligo dT primers (dilute to 10 µM; #18418012 ThermoFisher Scientific)
Random hexamer primers (dilute to 20 µM; #N8080127 ThermoFisher Scientific)
RNase inhibitor (dilute to 10 units/µl; #N211 Promega)
RNA Extraction/Purification Procedure:
- Collect up to 100 islets in an Eppendorf tube and add 350 µl RLT buffer* to disrupt cells.
- Vortex thoroughly and add to QIAShredder column with collection tube attached. Spin for 2 min at full speed in microcentrifuge to homogenize the sample.
- Add 350 µl 70% ethanol to the lysate and pipette repeatedly to mix.
- Transfer to an RNeasy spin column and spin at ≥8000xg for 15 secs to bind the RNA to the column. Discard the flow-through.
- Wash the column with 700 µl RW1 buffer and spin at ≥8000xg for 15 secs. Discard the flow-through.
- Wash the column with 500 µl RPE buffer* and spin at ≥8000xg for 15 secs. Discard the flow-through.
- Repeat column wash with 500 µl RPE buffer and spin at ≥8000xg for 2 min. Discard the flow-through.
- To elute the bound RNA, transfer column into a fresh Eppendorf tube and add 30 µl RNase-free water to membrane. Spin at ≥8000xg for 1 min.
- To ensure a high RNA concentration, use the eluate to repeat the elution by reapplying to the membrane and spinning at ≥8000xg for 1 min.*
- Determine the concentration and quality of the RNA sample.*
- To reverse transcribe template RNA into cDNA, prepare the following reaction in one eppendorf:
Up to 2 µg RNA*
4 µl of 10X Buffer RT
2 µl Oligo dT primer
2 µl random hexamer primer
1 µl RNase inhibitor
1 µl Reverse Transcriptase
Add RNase-free water to take reaction volume to 40 µl. - Mix thoroughly by briefly vortexing and centrifuge briefly to collect reaction to the bottom of the tube
- Incubate at 37oC for 60 min.
- Place reactions on ice and use up to 1 µl per 10 µl PCR reaction*.
Notes for this RNA Isolation Procedure:
- Step 1. On the day of RNA preparation, add 10 µl of 14.3 M 2-mercaptoethanol per 1ml RLT buffer prior to use. This helps disable RNAses.
- Step 6. Ensure ethanol is used to dilute RPE buffer concentrate (provided in kit) prior to first use
- Step 9. Store RNA samples at -80°C if required to prevent degradation
- Step 10. RNA analysis chip techniques such as Experion (BioRad) use a small amount of sample to provide accurate quantification and assessment of RNA quality
- Step 11. If RNA has been frozen, thaw on ice to avoid RNA degradation
- Step 14. Reactions can be stored at -20oC prior to PCR if required
Notes from our readers:
Mr. Young on Google + states:
1. [It is important to] …keep… an amplicon-free, RSase- free, clean environment
2. [Also have] a dedicated space for nucleic acid extraction separate from post amp and Master mix prep
Dr. Carina Jorgensen from Linked In states:
[Check the] integrity of … [your] … RNA … – that is whether or not its degraded – before …[you]…continue with downstream applications…You need to know the quality of RNA to choose your primer for cDNA synthesis, and if you want to continue with real-time qPCR you need to have a certain minimum level when it comes to the quality of your RNA, if you want to be sure that you can trust the following qPCR results. If you want to perform microchip analysis the demand to quality is even higher comparer to qPCR.
Guides for Specific Applications of RNA Extraction on Scigine
HeLA cell RNA Isolation
RNA Extraction from Cumulus Cells
RNA Extraction followed by RT-PCR
RNA Isolation and cDNA Synthesis
Acid-Phenol Extraction of RNA
References
Sambrook, J., and Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual. 3rd Ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
RNA Extraction Methods in Molecular Biology Book Chapter
Bitesize Bio Guide
[…] It might be useful to look at our RNA Extraction & Isolation guide if you’re planning on making cDNA related to your […]